Amino acids are organic compounds containing amine (-NH2) and carboxyl (-COOH) functional groups, along with a side chain which is specific to each amino acid. About 500 naturally occurring amino acids are known; only 20 appear in the genetic code (Tab. 1; Fig. 1). Amino acids can be classified according to their physical and chemical properties.
Tab. 1 Amino Acids; terms and abbreviation
| Amino acid | Short | Abbrev. | Amino acid | Short | Abbrev. |
| Alanine | A | Ala | Asparagine | N | Asn |
| Cysteine | C | Cys | Pyrrolysine | O | Pyl |
| Aspartic acid | D | Asp | Proline | P | Pro |
| Glutamic acid | E | Glu | Glutamine | Q | Gln |
| Phenylalanine | F | Phe | Arginine | R | Arg |
| Glycine | G | Gly | Serine | S | Ser |
| Histidine | H | His | Threonine | T | Thr |
| Isoleucine | I | Ile | Selenocysteine | U | Sec |
| Lysine | K | Lys | Valine | V | Val |
| Leucine | L | Leu | Tryptophan | W | Trp |
| Methionine | M | Met | Tyrosine | Y | Tyr |
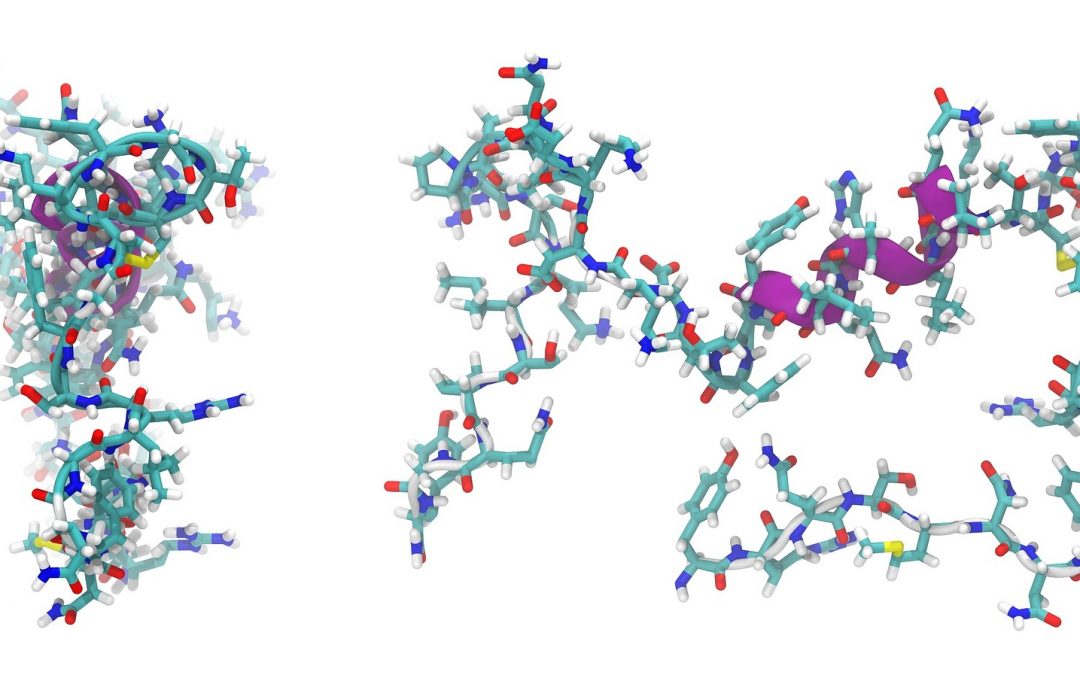
Proteins and peptides consist of one or more long chains of amino acid residues. Since they perform a variety of functions within organisms, they are of fundamental importance in our diet. Proteins and peptides are found in food, supplements chemical properties and cosmetics. Commonly, proteins are analyzed by liquid chromatography such as RP-HPLC or IC requiring high chemical amount, long hydrolysis as well as long analysis times. To overcome this challenge, new NMR methods are developed to identify and quantify amino acids in their free and bound form.
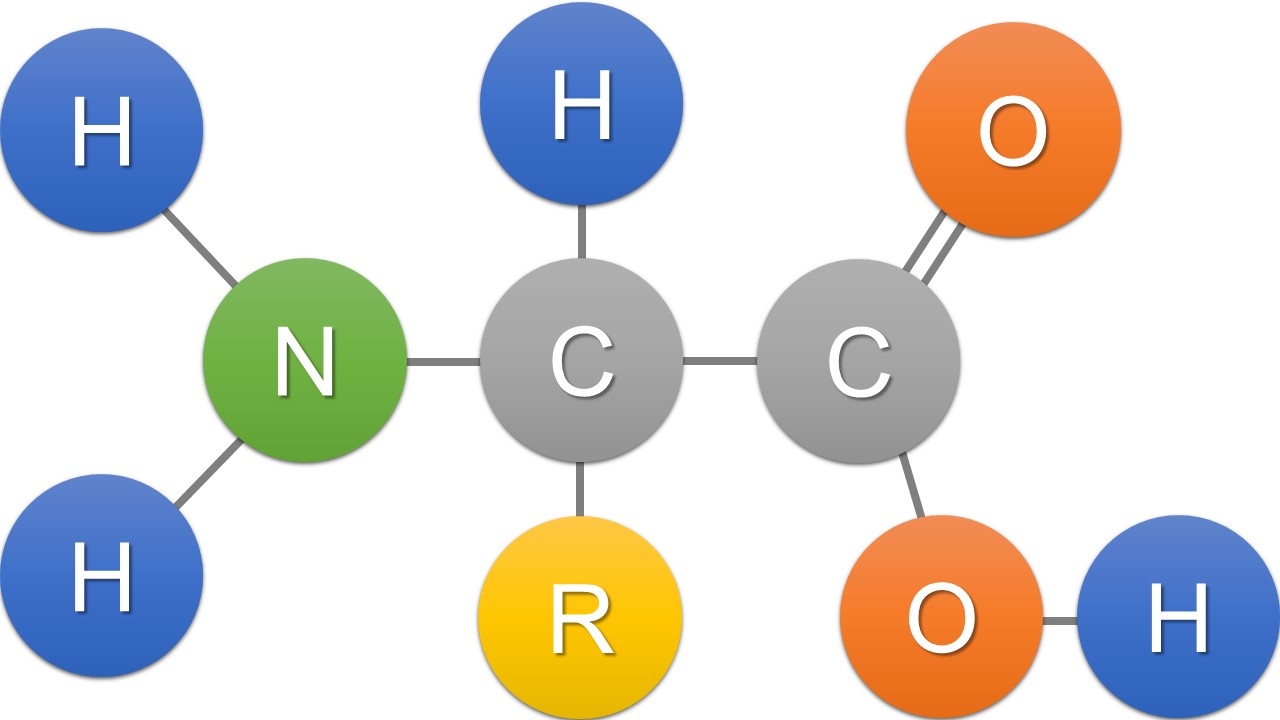
Fig. 1 The structure of an alpha amino acid in its un-ionized form
Sample preparation
For the amino acid analysis an appropriate amount of the sample is hydrolyzed in DCl. Evaporate the solvent and dissolves the residue in D2O. Transfer an appropriate amount of the solvent into an NMR tube. Use for the analysis a standard 13C NMR pulse sequence.
Results
NMR spectroscopy presents considerable information about the peptide structure. 1H NMR spectra are suitable for proofing the presence or absence of individual amino acids. To determine the amino acid sequence the peptide is analyzed by 2D-NMR experiments such as COSY, TOCSY, NOESY, HSQC and HMBC. Amino acids can be analyzed by 1H and 13C NMR spectroscopy after hydrolysis with DCl. After the acid hydrolysis, amino acids exist in their free form which can be analyzed by 1H and 13C NMR spectroscopy (Fig. 2). Their carbonyl signals appear in 13C NMR spectra between 169 ppm and 173 ppm. Each signal indicates one amino acid. Since the integral area is directly proportional to the molar amount, the amount of each amino acid can be determined quantitatively. Therefore, signal intensities are determined by automate deconvolution. Since the sum of all carbonyl signals is 100%, the relative intensities can be calculated with the respective molecular weights. The signal assignment of an exemplary protein is shown in Fig. 2. By this method 17 of the 20 natural amino acids can be investigated.
Since during hydrolysis Glutamine (Gln) is transformed into Glutamic acid (Glu) and Asparagine (Asn) into Aspartic acid (Asp), these amino acids are determined as a sum of [Gln + Glu] and [Asn + Asp]. Furthermore, Tryptophan (Trp) is generally not recovered under acid hydrolysis. Nevertheless, 17 amino acids can be determined in one single rapid 13C NMR measurement with a fast sample preparation.
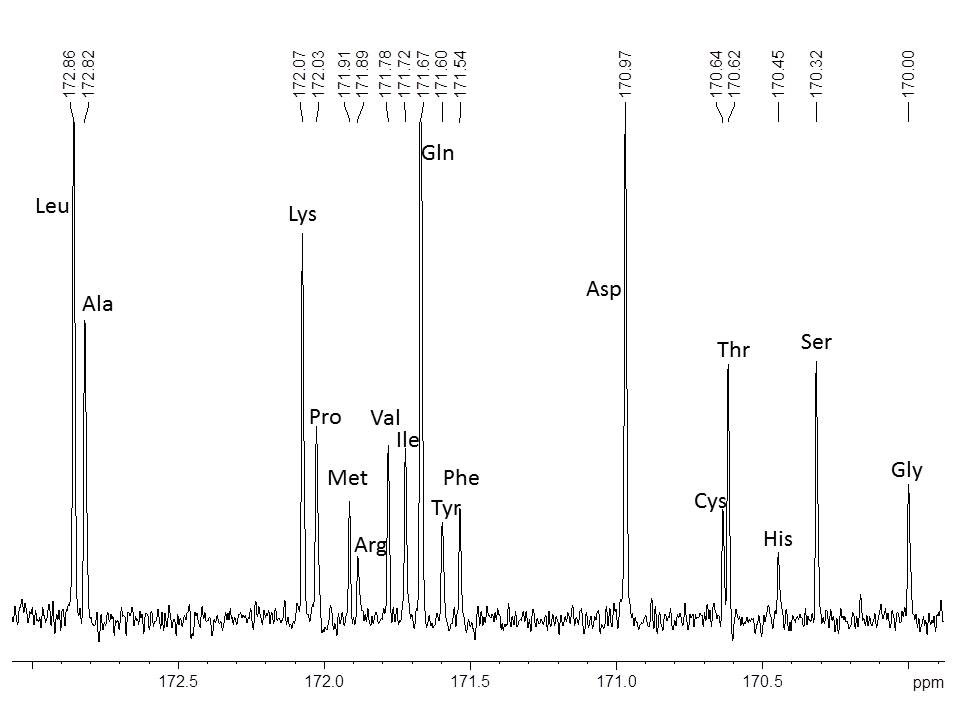
Fig. 2 13C NMR signal assignment of amino acids